Column guessing
How do you read the number 1.5? Ask Question Asked 4 years, 11 months ago. Active 4 years, 11 months ago. Viewed 6k times 11. How should you read the number 1.5? Should I read it as. One and half;. One and a half; Any help would be appreciated. Articles mathematics reading-aloud numbers.
The process by which readr guesses the types of columns has received a substantial overhaul to make it easier to fix problems when the initial guesses aren’t correct, and to make it easier to generate reproducible code. Now column specifications are printing by default when you read from a file:
And you can extract those values after the fact with spec():
This makes it easier to quickly identify parsing problems and fix them (#314). If the column specification is long, the new cols_condense() is used to condense the spec by identifying the most common type and setting it as the default. This is particularly useful when only a handful of columns have a different type (#466).
You can also generating an initial specification without parsing the file using spec_csv(), spec_tsv(), etc.
Once you have figured out the correct column types for a file, it’s often useful to make the parsing strict. You can do this either by copying and pasting the printed output, or for very long specs, saving the spec to disk with write_rds(). In production scripts, combine this with stop_for_problems() (#465): if the input data changes form, you’ll fail fast with an error.
You can now also adjust the number of rows that readr uses to guess the column types with guess_max:
You can now access the guessing algorithm from R. guess_parser() will tell you which parser readr will select for a character vector (#377). We’ve made a number of fixes to the guessing algorithm:
New example
extdata/challenge.csvwhich is carefully created to cause problems with the default column type guessing heuristics.Blank lines and lines with only comments are now skipped automatically without warning (#381, #321).
Single ‘-’ or ‘.’ are now parsed as characters, not numbers (#297).
Numbers followed by a single trailing character are parsed as character, not numbers (#316).
We now guess at times using the
time_formatspecified in thelocale().
We have made a number of improvements to the reification of the col_types, col_names and the actual data:
If
col_typesis too long, it is subsetted correctly (#372, @jennybc).If
col_namesis too short, the added names are numbered correctly (#374, @jennybc).Missing colum name names are now given a default name (
X2,X7etc) (#318). Duplicated column names are now deduplicated. Both changes generate a warning; to suppress it supply an explicitcol_names(settingskip = 1if there’s an existing ill-formed header).col_types()accepts a named list as input (#401).
- Reading a file
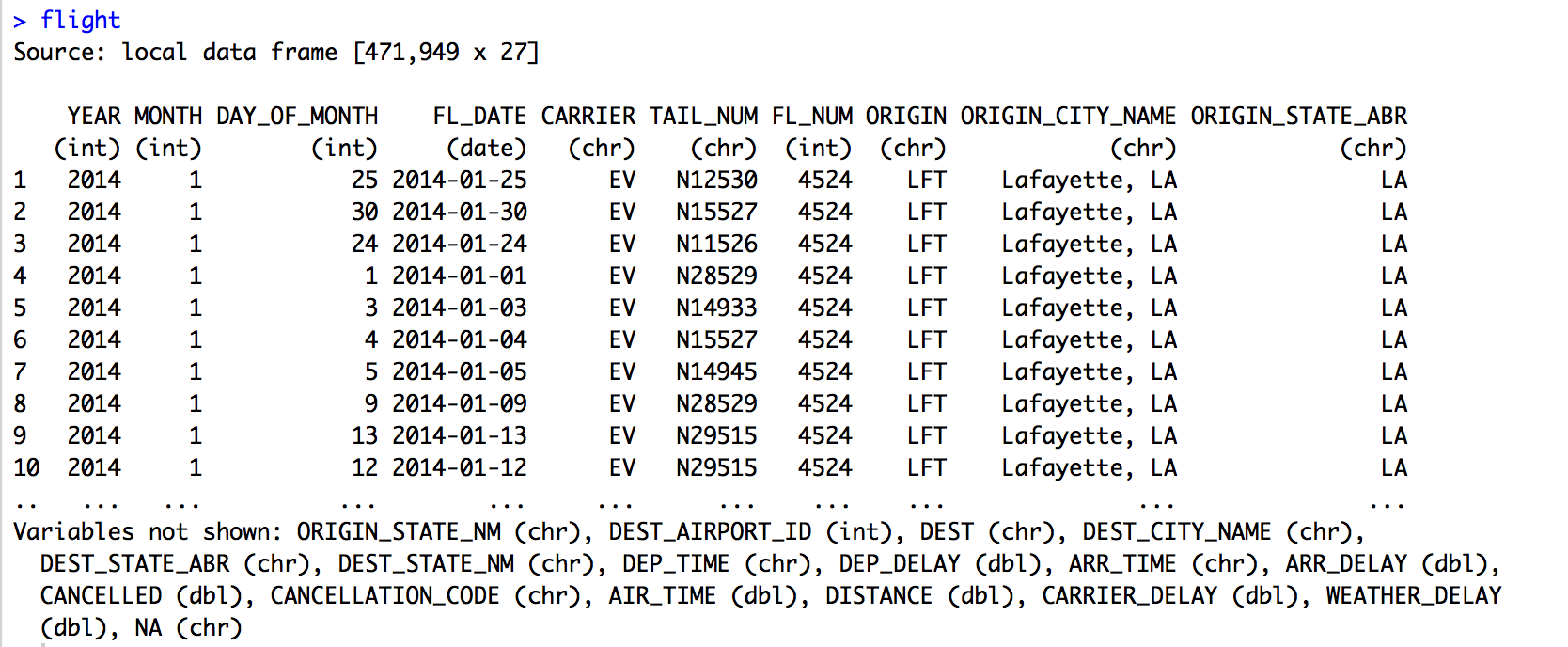
There are many solutions for importingtxt|csv file into R. In our previous articles, we described some best practices for preparing your data as well as R base functions (read.delim() and read.csv()) for importing txt|csv file into R.
In this article, we’ll describe the readr package, developed by Hadley Wickham. readr package provides a fast and friendly solution to read a delimited file into R.
Compared to R base functions, readr functions are:
- much faster (X10),
- have a helpful progress bar if loading is going to take a while and
- all functions work exactly the same way.
Launch RStudio as described here: Running RStudio and setting up your working directory
Prepare your data as described here: Best practices for preparing your data
The readr package contains functions for reading i) delimited files, ii) lines and iii) the whole file.
The function read_delim()[in readr package] is a general function to import a data table into R. Depending on the format of your file, you can also use:

- read_csv(): to read a comma (“,”) separated values
- read_csv2(): to read a semicolon (“;”) separated values
- read_tsv(): to read a tab separated (“t”) values
The simplified format of these functions are, as follow:
- file: file path, connexion or raw vector. Files ending in .gz, .bz2, .xz, or .zip will be automatically uncompressed. Files starting with “http://”, “https://”, “ftp://”, or “ftps://” will be automatically downloaded. Remote gz files can also be automatically downloaded & decompressed.
- delim: the character that separates the values in the data file.
- col_names: Either TRUE, FALSE or a character vector specifying column names. If TRUE, the first row of the input will be used as the column names.
read_csv() and read_tsv() are special case of the general function read_delim(). They’re useful for reading the most common types of flat file data, comma separated values and tab separated values, respectively.
The above mentioned functions return an object of class tbl_df which is data frame providing a nicer printing method, useful when working with large data sets.
Reading a local file
- To import a local .txt or .csv files, the syntax would be:
The above R code, assumes that the file “mtcars.txt” or “mtcars.csv” is in your current working directory. To know your current working directory, type the function getwd() in R console.

- It’s also possible to choose a file interactively using the function file.choose(), which I recommend if you’re a beginner in R programming:
If you use the R code above in RStudio, you will be asked to choose a file.
- If your field separator is for example “|”, it’s possible to use the general function read_delim(), which reads in files with a user supplied delimiter:
Reading a file from internet
It’s possible to use the functions read_delim(), read_csv() and read_tsv() to import files from the web.
In the case of parsing problems
If there are parsing problems, a warning tells you how many, and you can retrieve the details with the function problems().
There are different types of data: numeric, character, logical, …
readr tries to guess automatically the type of data contained in each column. You might see a lot of warnings in a situation where readr has guessed the column type incorrectly. To fix these problems you can use the additional arguments col_type() to specify the data type of each column.
The following column types are available:
- col_integer(): to specify integer (alias = “i”)
- col_double(): to specify double (alias = “d”).
- col_logical(): to specify logical variable (alias = “l”)
- col_character(): leaves strings as is. Don’t convert it to a factor (alias = “c”).
- col_factor(): to specify a factor (or grouping) variable (alias = “f”)
- col_skip(): to ignore a column (alias = “-” or “_“)
- col_date() (alias = “D”), col_datetime() (alias = “T”) and col_time() (“t”) to specify dates, date times, and times.
An example is as follow (column x is an integer (i) and column treatment = “character” (c):
Function: read_lines().
Readers 1.50 Glasses
- Simplified format:
- file: file path
- skip: Number of lines to skip before reading data
- n_max: Numbers of lines to read. If n is -1, all lines in file will be read.
The function read_lines() returns a character vector with one element for each line.
- Example of usage
Import a local .txt file: read_tsv(file.choose())
Import a local .csv file: read_csv(file.choose())
- Import a file from internet: read_delim(url) if a txt file or read_csv(url) if a csv file
- Previous chapters
- Next chapters
This analysis has been performed using R (ver. 3.2.3).
Show me some love with the like buttons below... Thank you and please don't forget to share and comment below!!
Montrez-moi un peu d'amour avec les like ci-dessous ... Merci et n'oubliez pas, s'il vous plaît, de partager et de commenter ci-dessous!
Recommended for You!
More books on R and data science
Want to Learn More on R Programming and Data Science?
Follow us by EmailOn Social Networks:
Readers 1.5
Readers 1.50 2.oo. 2.50
Click to follow us on Facebook and Google+ :
Comment this article by clicking on 'Discussion' button (top-right position of this page)